Nearly all proteins have structural similarities
with other proteins and, in many cases, share a
common evolutionary origin. The knowledge of
these relationships makes important contributions to
molecular biology and to other related areas of
science. It is central to our understanding of the
structure and evolution of proteins. It will play an
important role in the interpretation of the sequences
produced by the genome projects and, therefore, in
understanding the evolution of development.
The recent exponential growth in the number of
proteins whose structures have been determined by
X-ray crystallography and NMR spectroscopy
means that there is now a large and rapidly growing
corpus of information available. At present (January,
1995) the Brookhaven Protein Databank (PDB,
(Abola et al., 1987)) contains 3091 entries and the
number is increasing by about 100 a month. To
facilitate the understanding of, and access to, this
information, we have constructed the Structural
Classification of Proteins (scop) database. This
database provides a detailed and comprehensive
description of the structural and evolutionary
relationships of proteins whose three-dimensional
structures have been determined. It includes all proteins in the current version of the PDB and
almost all proteins for which structures have been
published but whose co-ordinates are not available
from the PDB.
The classification of protein structures in the
database is based on evolutionary relationships and
on the principles that govern their three-dimensional
structure. Early work on protein structures showed
that there are striking regularities in the ways in
which secondary structures are assembled (Levitt
& Chothia, 1976; Chothia et al., 1977) and in the
topologies of the polypeptide chains (Richardson,
1976, 1977; Sternberg & Thornton, 1976). These
regularities arise from the intrinsic physical and
chemical properties of proteins (Chothia, 1984;
Finkelstein&Ptitsyn, 1987) and provide the basis for
the classification of protein folds (Levitt & Chothia,
1976; Richardson, 1981). This early work has been
taken further inmore recent papers; see, for example,
Holm & Sander (1993), Orengo et al. (1993),
Overington et al. (1993) and Yee & Dill (1993). An
extensive bibliography of papers on the classification
and the determinants of protein folds is given in scop.
The method used to construct the protein
classification in scop is essentially the visual
inspection and comparison of structures though
various automatic tools are used to make the task
manageable and help provide generality. Given the current limitations of purely automatic procedures,
we believe this approach produces the most
accurate and useful results. The unit of classification
is usually the protein domain. Small
proteins, and most of those of medium size, have
a single domain and are, therefore, treated as a
whole. The domains in large proteins are usually
classified individually.
The classification is on hierarchical levels that
embody the evolutionary and structural relationships.
FAMILY. Proteins are clustered together into
families on the basis of one of two criteria that imply
their having a common evolutionary origin: first, all
proteins that have residue identities of 30% and
greater; second, proteins with lower sequence identities but whose functions and structures are
very similar; for example, globins with sequence
identities of 15%.
SUPERFAMILY. Families, whose proteins have
low sequence identities but whose structures and, in
many cases, functional features suggest that a
common evolutionary origin is probable, are placed
together in superfamilies; for example, actin, the
ATPase domain of the heat-shock protein and
hexokinase (Flaherty et al., 1991).
COMMONFOLD. Superfamilies and families are
defined as having a common fold if their proteins
have same major secondary structures in same
arrangement with the same topological connections.
In scop we give for each fold short descriptions of its
main structural features. Different proteins with the
same fold usually have peripheral elements of
secondary structure and turn regions that differ in
size and conformation and, in the more divergent
cases, these differing regions may form half or more
of each structure. For proteins placed together in the
same fold category, the structural similarities
probably arise from the physics and chemistry of
proteins favouring certain packing arrangements and
chain topologies (see above). There may, however,
be cases where a common evolutionary origin is
obscured by the extent of the divergence in sequence,
structure and function. In these cases, it is possible
that the discovery of new structures, with folds
between those of the previously known structures,
will make clear their common evolutionary relationship.
CLASS. For convenience of users, the different
folds have been grouped into classes. Most of the
folds are assigned to one of the five structural classes
on the basis of the secondary structures of which
they composed: (1) all alpha (for proteins whose
structure is essentially formed by a-helices), (2) all
beta (for those whose structure is essentially formed
by b-sheets), (3) alpha and beta (for proteins with
a-helices and b-strands that are largely interspersed),
(4) alpha plus beta (for those in which
a-helices and b-strands are largely segregated) and
(5)multi-domain (for those with domains of different
fold and for which no homologues are known at
present). Note that we do not use Greek characters
in scop because they are not accessible to all world
wide web viewers. More unusual proteins, peptides
and the PDB entries for designed proteins theoretical models, nucleic acids and carbohydrates,
have been assigned to other classes.
The number of entries, families, superfamilies and
common folds in the current version of scop are
shown in Figure 1. The exact position of boundaries
between family, superfamily and fold are, to some
degree, subjective. However, because all proteins
that could conceivably belong to a family or
superfamily are clustered together in the encompassing
fold category, some users may wish to
concentrate on this part of the database.
In addition to the information on structural and
evolutionary relationships, each entry (for which
co-ordinates are available) has links to images of the
structure, interactive molecular viewers, the atomic
co-ordinates, sequence data and homologues and
MEDLINE abstracts (see Table 1).
Two search facilities are available in scop. The
homology search permits users to enter a sequence
and obtain a list of any structures to which it has
significant levels of sequence similarity. The key
word search finds, for a word entered by the user,
matches from both the text of the scop database and
the headers of Brookhaven Protein Databank
structure files.
To provide easy and broad access, we have made
the scop database available as a set of tightly coupled
hypertext pages on the world wide web (WWW).
This allows it to be accessed by any machine on the
internet (including Macintoshes, PCs and workstations)
using freeWWWreader programs, such as
Mosaic (Schatz & Hardin, 1994). Once such a
program has been started, it is necessary only to
‘‘open’’ URL:
http://scop.mrc-lmb.cam.ac.uk/scop/
to obtain the ‘‘home’’ page level of the database.
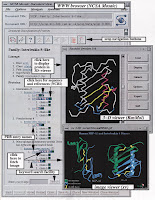
In Figure 2 we show a typical page from the
database. Each page has buttons to go back to the
top-level home page, to send electronic mail to the
authors, and to retrieve a detailed help page.
Navigating through the tree structure is simple;
selecting any entry retrieves the appropriate page. In
addition, buttons make it possible tomove within the
hierarchy in other manners, such as ‘‘upwards’’ to
obtain broader levels of classification.
The scop database was originally created as a
tool for understanding protein evolution through
sequence-structure relationships and determining if
new sequences and new structures are related to
previously known protein structures. On a more
general level, the highest levels of classification
provide an overview of the diversity of protein
structures now known and would be appropriate
both for researchers and students. The specific lower
levels should be helpful for comparing individual
structures with their evolutionary and structurally
related counterparts. In addition, we have also found
that the search capabilities with easy access to data
and images make scop a powerful general-purpose
interface to the PDB.
As new structures are released by PDB and
published, they will be entered in scop and revised versions of the database will be made available on
WWW. Moreover, as our formal understanding of
relationships between structure, sequence function
and evolution grows, it will be embodied in
additional facilities in the database.
No comments:
Post a Comment