Wednesday, April 8, 2009
Protein Databases
A database is a collection of similar information which is stored in the computer system.
In case of Bioinformatics, Databases are developed for Drug designing, Clinical Data or any simple information on Proteins, Nucleotides, Genes, Gene prediction and so on.
Can be created by anyone who has good computer knowledge.
Protein Database
Collection of similar protein information: Sequence or Structure.
The three being discussed today:-
-> PDB
-> dbPTM
-> SCOP
Protein Data Bank (PDB)
Belongs to the RCSB (Research Collaboratory for Structural Bioinformatics).
A repository for the 3-D structural data of large biological molecules, proteins and nucleic acids.
The PDB is a key resource in areas of structural biology, such as structural genomics.
Data obtained by X-ray crystallography or NMR spectroscopy.
Overseen by an organization called the Worldwide Protein Data Bank.
The PDB database is updated every Tuesday.
The PDB ID is of four characters- first is any number from 1 to 9, rest can be alpha-numeric.
In 2007, 7263 structures were added. In 2008, only 7073 structures were added, with a total of 55,660 structures.
The information summarized for each entry includes several data items:
Title- The title of the experiment or analysis that is represented in the entry.
Author- The names of the authors responsible for the deposition.
Primary Citation- Includes the primary journal reference to the structure.
History- Includes the date of deposition, date of release of the structure by PDB and supersedes (appears if a previous version, or versions, of a structure were deposited with the PDB.
Experimental Method- The experimental technique used to solve the structure including theoretical modeling.
Parametres- For structures that were determined by x-ray diffraction, this section gives information about the refinement of the structure.
Unit Cell- For structures that were determined by x-ray diffraction, this section gives the crystal cell lengths and angles.
NMR Ensemble-For structures determined by NMR, this section includes the total number of conformer that were calculated in the final round, number of conformer that are submitted for the ensemble & description of how the submitted conformer (models) were selected.
NMR Refine- Contains the method used to determine the structure.
Molecular Description- Contains the no. of polymers, molecule name, any mutation, if present, entity fragment description, chain identifiers and the EC (Enzyme Commission) number.
Source- Specifies the biological and/or chemical source of the molecule given for each entity identified in the molecular description section.
Related PDB entries- Data items in this section contain references to entries that are related to the entry.
Chemical Component- Contains the 3-letter code, the name & the chemical formula of the chemical component.
SCOP classification- Classifications are pulled from the SCOP database and summarized here.
CATH classification- As classified by the CATH database.
GO Terms- Clicking on any of the results in this section will perform a search of the database resulting in a Query Results Browser page containing all structures with the selected Molecular Function, Biological Process Cellular Component.
dbPTM
dbPTM is a database that compiles information on protein post-translational modifications (PTMs), such as the catalytic sites, solvent accessibility of amino acid residues, protein secondary and tertiary structures, protein domains and protein variations.
The database includes all the experimentally validated PTM sites from Swiss-Prot, PhosphoELM and O-GLYCBASE.
The dbPTM systematically identifies three major types of protein PTM (phosphorylation, glycosylation and sulfation) sites against Swiss-Prot proteins.
The summary table of PTMs :
To facilitate the users to investigate and browse all the types of PTM in the release 2.0 of dbPTM. In the table, each type of PTM was categorized by their modified amino acids with the number of experimentally verified sites. For example, users can choose the acetylation of lysine (K) to take the more detailed information such as the position of modification on amino acid, the location of modification on protein sequence, the modified chemical formula, and the mass difference.The most effective knowledge about the PTM is the substrate site specificity including the frequency of amino acids, the average solvent accessibility, and the frequency of secondary structure surrounding the modified site.
Wednesday, April 1, 2009
Clinical Trials
Clinical Trials
Clinical trials: Trials to evaluate the effectiveness and safety of medications or medical devices by monitoring their effects on large groups of people.
Clinical research trials may be conducted by government health agencies such as NIH, researchers affiliated with a hospital or university medical program, independent researchers, or private industry.
Depending on the type of product and the stage of its development, investigators enroll healthy volunteers and/or patients into small pilot studies initially, followed by larger scale studies in patients that often compare the new product with the currently prescribed treatment. As positive safety and efficacy data are gathered, the number of patients is typically increased. Clinical trials can vary in size from a single center in one country to multicenter trials in multiple countries.
Usually volunteers are recruited, although in some cases research subjects may be paid. Subjects are generally divided into two or more groups, including a control group that does not receive the experimental treatment, receives a placebo (inactive substance) instead, or receives a tried-and-true therapy for comparison purposes.
Typically, government agencies approve or disapprove new treatments based on clinical trial results. While important and highly effective in preventing obviously harmful treatments from coming to market, clinical research trials are not always perfect in discovering all side effects, particularly effects associated with long-term use and interactions between experimental drugs and other medications.
For some patients, clinical research trials represent an avenue for receiving promising new therapies that would not otherwise be available. Patients with difficult to treat or currently "incurable" diseases, such as AIDS or certain types of cancer, may want to pursue participation in clinical research trials if standard therapies are not effective. Clinical research trials are sometimes lifesaving.
There are four possible outcomes from a clinical trial:
Positive trial -- The clinical trial shows that the new treatment has a large beneficial effect and is superior to standard treatment.
Non-inferior trial -- The clinical trial shows that that the new treatment is equivalent to standard treatment. Also called a non-inferiority trial.
Inconclusive trial -- The clinical trial shows that the new treatment is neither clearly superior nor clearly inferior to standard treatment.
Negative trial -- The clinical trial shows that a new treatment is inferior to standard treatment.
History
Clinical trials were first introduced in Avicenna's The Canon of Medicine in 1025 AD, in which he laid down rules for the experimental use and testing of drugs and wrote a precise guide for practical experimentation in the process of discovering and proving the effectiveness of medical drugs and substances. He laid out the following rules and principles for testing the effectiveness of new drugs and medications, which still form the basis of modern clinical trials:
1."The drug must be free from any extraneous accidental quality."
2."It must be used on a simple, not a composite, disease."
3."The drug must be tested with two contrary types of diseases, because sometimes a drug cures one disease by its essential qualities and another by its accidental ones."
4."The quality of the drug must correspond to the strength of the disease. For example, there are some drugs whose heat is less than the coldness of certain diseases, so that they would have no effect on them."
5."The time of action must be observed, so that essence and accident are not confused."
6."The effect of the drug must be seen to occur constantly or in many cases, for if this did not happen, it was an accidental effect."
7."The experimentation must be done with the human body, for testing a drug on a lion or a horse might not prove anything about its effect on man."
One of the most famous clinical trials was James Lind's demonstration in 1747 that citrus fruits cure scurvy. He compared the effects of various different acidic substances, ranging from vinegar to cider, on groups of afflicted sailors, and found that the group who were given oranges and lemons had largely recovered from scurvy after 6 days.
Possible advantages
Clinical trials are done with the sole aim of testing medicines, medical devices and treatments that will ultimately be made available for human health. By participating in trials:
You may gain access during and after the clinical trial to new treatments that are not yet available to the general population
You may obtain medical care free of charge
You will be closely monitored for possible adverse events
You are contributing to medical research that may result in the advancement of medicine and healthcare in general thereby helping other fellow human beings
Participating in clinical trials is not a source of primary or additional income. However almost all sponsors reimburse persons that participate in trials for all reasonable expenses related to participating in the trial, including travel expenses, food, medical care and compensation for provable and insured adverse events that are related to the trial.
Possible disadvantages
There may be serious adverse events (SAEs) that are related to the medications used or procedures that are done in the trial; however study participants are intensively monitored so that SAEs may be detected early and managed appropriately. There is also insurance cover for some SAEs, so that participants may be compensated accordingly.
The medicines, vaccines, medical devices or treatment options used in the trial may not be effective for your disease; there are, however, safety procedures in place so that those participants who do not benefit from the trial medical management options may be switched to alternative effective treatment immediately or at the end of the trial.
The trial guidelines may require some adjustment of one or more aspects of your life, such as:
You may need to set aside time for trial related activities like visiting the trial site
You may not use certain medications including traditional medications without the approval of your trial doctor
Your personal private or social life may be affected, e.g. sexual activity, reproductive functioning, consumption of alcohol, tobacco or other drugs of abuse, etc.
You may have to consult your usual healthcare provider for all your other illnesses that are not related to the trial, but still you have to inform your provider that you are part of a trial and that certain medications or treatment options may not be compatible with your trial protocol
Your employer, medical aid, personal insurance and/or Commissioner for Compensation for Occupational Injuries may not pay for claims that are related to events due to your participation in clinical trials; it is therefore extremely important that you verify that the sponsor of the trial has an appropriate comprehensive insurance cover for you.
Design Of Clinical Trials
A fundamental distinction in evidence-based medicine is between observational studies and randomized controlled trials. Types of observational studies in epidemiology such as the cohort study and the case-control study provide less compelling evidence than the randomized controlled trial. In observational studies, the investigators only observe associations (correlations) between the treatments experienced by participants and their health status or diseases.
A randomized controlled trial is the study design that can provide the most compelling evidence that the study treatment causes the expected effect on human health.
Currently, some Phase II and most Phase III drug trials are designed as randomized, double blind, and placebo-controlled.
Randomized: Each study subject is randomly assigned to receive either the study treatment or a placebo.
Blind: The subjects involved in the study do not know which study treatment they receive. If the study is double-blind, the researchers also do not know which treatment is being given to any given subject. This 'blinding' is to prevent biases, since if a physician knew which patient was getting the study treatment and which patient was getting the placebo, he/she might be tempted to give the (presumably helpful) study drug to a patient who could more easily benefit from it. In addition, a physician might give extra care to only the patients who receive the placebos to compensate for their ineffectiveness. A form of double-blind study called a "double-dummy" design allows additional insurance against bias or placebo effect. In this kind of study, all patients are given both placebo and active doses in alternating periods of time during the study.
Placebo-controlled: The use of a placebo (fake treatment) allows the researchers to isolate the effect of the study treatment.
Although the term "clinical trials" is most commonly associated with the large, randomized studies typical of Phase III, many clinical trials are small. They may be "sponsored" by single physicians or a small group of physicians, and are designed to test simple questions. In the field of rare diseases sometimes the number of patients might be the limiting factor for a clinical trial. Other clinical trials require large numbers of participants (who may be followed over long periods of time), and the trial sponsor is a private company, a government health agency, or an academic research body such as a university.
Clinical Trial Design — What do probability and statistics have to do with it?
People are familiar with the idea of random variability. When you flip a coin 10 times, you “expect” 5 heads and 5 tails—but you’re not at all surprised to get different numbers. Perhaps this time you might get 6 and 4 . . . or perhaps 4 and 6. You could get 7 and 3, and you wouldn’t be knocked out of your chair if you got 8 and 2.
In fact, the chance of a theoretically “perfect” 5 and 5 outcome is only 24.6%. In other words, if 100 people try flipping a coin 10 times, only about 25 of them would see the “correct” ratio.
The chance discussed above—of getting 8 heads—has 4.4% probability. Since the chance of 8 tails is the same, their combined probability is 4.4% + 4.4% = 8.8%. So, out of 100 people, we’d expect about 9 to get either 8 heads and 2 tails or else 2 tails and 8 heads. In practice, lopsided outcomes are a definite, if infrequent, occurrence: some observers will get them. The probability of all heads is only 0.1% so, in a hundred people, we likely wouldn’t see anybody getting that; but in a crowd of 1000 people there could be 1 with all heads—and another with all tails.
In clinical trials the variation arises because the random selection of subjects and their random assignment to treatment could bring an atypically large number of “difficult” or of “easy” subjects to one treatment over the other. Treatment A, which has a true success rate of 50%, could easily show 3 successes in 10 subjects, while Treatment B, which has a true rate of only 40%, could show 5 successes in 10 subjects. Then, based on our total combined sample of 20, we could become wrongly, stubbornly convinced that Treatment B is better.
The squares in the gray-shaded diagonal represent outcomes in which an equal number of successes are observed for both treatments, even though Treatment A is superior. The sum total of the probabilities of these outcomes equals 16.0%. In practical terms, if this were an experiment assigned by a biology professor to a class of 100 students, it could be expected that 16 students would get data wrongly suggesting that the two treatments are equally effective. The black-shaded squares above the diagonal represent outcomes in which Treatment B is observed to have more successes than Treatment A. The combined probability of these outcomes equals 24.8%, so the biology professor can expect about 25 of the 100 students to submit lab reports concluding wrongly that Treatment B is superior. Only 59 students in the class of 100 will observe data (the white shaded squares below the diagonal) that will lead them to the correct conclusion.
Clinical trial protocol
A clinical trial protocol is a document used to gain confirmation of the trial design by a panel of experts and adherence by all study investigators, even if conducted in various countries.
The protocol describes the scientific rationale, objective(s), design, methodology, statistical considerations, and organization of the planned trial. Details of the trial are also provided in other documents referenced in the protocol such as an Investigator's Brochure.
The protocol contains a precise study plan for executing the clinical trial, not only to assure safety and health of the trial subjects, but also to provide an exact template for trial conduct by investigators at multiple locations (in a "multicenter" trial) to perform the study in exactly the same way. This harmonization allows data to be combined collectively as though all investigators (referred to as "sites") were working closely together. The protocol also gives the study administrators (often a contract research organization) as well as the site team of physicians, nurses and clinic administrators a common reference document for site responsibilities during the trial.
The format and content of clinical trial protocols sponsored by pharmaceutical, biotechnology or medical device companies in the United States, European Union, or Japan has been standardized to follow Good Clinical Practice guidance[10] issued by the International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH).[11] Regulatory authorities in Canada and Australia also follow ICH guidelines. Some journals, e.g. Trials, encourage trialists to publish their protocols in the journal.
Design features
Informed consent
An essential component of initiating a clinical trial is to recruit study subjects following procedures using a signed document called "informed consent."[12]
Informed consent is a legally-defined process of a person being told about key facts involved in a clinical trial before deciding whether or not to participate. To fully describe participation to a candidate subject, the doctors and nurses involved in the trial explain the details of the study. Foreign language translation is provided if the participant's native language is not the same as the study protocol.
The research team provides an informed consent document that includes trial details, such as its purpose, duration, required procedures, risks, potential benefits and key contacts. The participant then decides whether or not to sign the document in agreement. Informed consent is not an immutable contract, as the participant can withdraw at any time.
Statistical power
In designing a clinical trial, a sponsor must decide on the target number of patients who will participate. The sponsor's goal usually is to obtain a statistically significant result showing a significant difference in outcome (e.g., number of deaths after 28 days in the study) between the groups of patients who receive the study treatments. The number of patients required to give a statistically significant result depends on the question the trial wants to answer. For example, to show the effectiveness of a new drug in a non-curable disease as metastatic kidney cancer requires many fewer patients than in a highly curable disease as seminoma if the drug is compared to a placebo.
The number of patients enrolled in a study has a large bearing on the ability of the study to reliably detect the size of the effect of the study intervention. This is described as the "power" of the trial. The larger the sample size or number of participants in the trial, the greater the statistical power.
However, in designing a clinical trial, this consideration must be balanced with the fact that more patients make for a more expensive trial. The power of a trial is not a single, unique value; it estimates the ability of a trial to detect a difference of a particular size (or larger) between the treated (tested drug/device) and control (placebo or standard treatment) groups. By example, a trial of a lipid-lowering drug versus placebo with 100 patients in each group might have a power of .90 to detect a difference between patients receiving study drug and patients receiving placebo of 10 mg/dL or more, but only have a power of .70 to detect a difference of 5 mg/dL.
Placebo groups
Merely giving a treatment can have nonspecific effects, and these are controlled for by the inclusion of a placebo group. Subjects in the treatment and placebo groups are assigned randomly and blinded as to which group they belong. Since researchers can behave differently to subjects given treatments or placebos, trials are also doubled-blinded so that the researchers do not know to which group a subject is assigned.
Assigning a person to a placebo group can pose an ethical problem if it violates his or her right to receive the best available treatment. The Declaration of Helsinki provides guidelines on this issue.
Phases of a Clinical Trial
Phase 0
Phase 0 is a recent designation for exploratory, first-in-human trials conducted in accordance with the U.S. Food and Drug Administration’s (FDA) 2006 Guidance on Exploratory Investigational New Drug (IND) Studies.Phase 0 trials are also known as human microdosing studies and are designed to speed up the development of promising drugs or imaging agents by establishing very early on whether the drug or agent behaves in human subjects as was expected from preclinical studies. Distinctive features of Phase 0 trials include the administration of single subtherapeutic doses of the study drug to a small number of subjects (10 to 15) to gather preliminary data on the agent's pharmacokinetics (how the body processes the drug) and pharmacodynamics (how the drug works in the body).
A Phase 0 study gives no data on safety or efficacy, being by definition a dose too low to cause any therapeutic effect. Drug development companies carry out Phase 0 studies to rank drug candidates in order to decide which has the best pharmacokinetic parameters in humans to take forward into further development. They enable go/no-go decisions to be based on relevant human models instead of relying on sometimes inconsistent animal data.
Phase 1
In Phase I a small number of healthy volunteers are exposed to the research treatment. The method of delivery as well as the dosing regimen is explored during this phase, and side effects are noted. Before Phase I studies begin, experiments comparing the new treatment with the drug of choice for the planned condition have been done in laboratory models and in animal studies, as well as extensive animal toxicity studies.
There are different kinds of Phase I trials:
SAD
Single Ascending Dose studies are those in which small groups of subjects are given a single dose of the drug while they are observed and tested for a period of time. If they do not exhibit any adverse side effects, and the pharmacokinetic data is roughly in line with predicted safe values, the dose is escalated, and a new group of subjects is then given a higher dose. This is continued until pre-calculated pharmacokinetic safety levels are reached, or intolerable side effects start showing up (at which point the drug is said to have reached the Maximum tolerated dose (MTD).
MAD
Multiple Ascending Dose studies are conducted to better understand the pharmacokinetics & pharmacodynamics of multiple doses of the drug. In these studies, a group of patients receives multiple low doses of the drug, whilst samples (of blood, and other fluids) are collected at various time points and analyzed to understand how the drug is processed within the body. The dose is subsequently escalated for further groups, up to a predetermined level.
Food effect
A short trial designed to investigate any differences in absorption of the drug by the body, caused by eating before the drug is given. These studies are usually run as a crossover study, with volunteers being given two identical doses of the drug on different occasions; one while fasted, and one after being fed.
Phase II
In Phase II, the effectiveness of the new treatment is characterized. The new drug is examined in patients using strict design criteria - appropriate monitoring, use of adequate controls, careful exploration of the effective and safe dose range, etc. Phase II studies are sometimes divided into Phase IIA and Phase IIB.
Phase IIA is specifically designed to assess dosing requirements (how much drug should be given).
Phase IIB is specifically designed to study efficacy (how well the drug works at the prescribed dose(s)).
Some trials combine Phase I and Phase II, and test both efficacy and toxicity.
Some Phase II trials are designed as case series, demonstrating a drug's safety and activity in a selected group of patients. Other Phase II trials are designed as randomized clinical trials, where some patients receive the drug/device and others receive placebo/standard treatment. Randomized Phase II trials have far fewer patients than randomized Phase III trials.
Phase III
In Phase III, large studies are done to compare the new medicament against a recognized standard treatment. Again, the studies must be well-controlled and well-conducted, to provide clear cut evidence of safety and effectiveness for the new drug regulatory authorities (e.g. FDA).
While not required in all cases, it is typically expected that there be at least two successful Phase III trials, demonstrating a drug's safety and efficacy, in order to obtain approval from the appropriate regulatory agencies such as FDA (USA), TGA (Australia), EMEA (European Union), or CDSCO/ICMR (India), for example.
Once a drug has proved satisfactory after Phase III trials, the trial results are usually combined into a large document containing a comprehensive description of the methods and results of human and animal studies, manufacturing procedures, formulation details, and shelf life. This collection of information makes up the "regulatory submission" that is provided for review to the appropriate regulatory authorities[1] in different countries. They will review the submission, and, it is hoped, give the sponsor approval to market the drug.
Phase IV
Phase IV trial is also known as Post Marketing Surveillance Trial. Phase IV trials involve the safety surveillance (pharmacovigilance) and ongoing technical support of a drug after it receives permission to be sold. Phase IV studies may be required by regulatory authorities or may be undertaken by the sponsoring company for competitive (finding a new market for the drug) or other reasons (for example, the drug may not have been tested for interactions with other drugs, or on certain population groups such as pregnant women, who are unlikely to subject themselves to trials). The safety surveillance is designed to detect any rare or long-term adverse effects over a much larger patient population and longer time period than was possible during the Phase I-III clinical trials. Harmful effects discovered by Phase IV trials may result in a drug being no longer sold, or restricted to certain uses: recent examples involve cerivastatin (brand names Baycol and Lipobay), troglitazone (Rezulin) and rofecoxib (Vioxx).
Ethical Conduct
Clinical trials are closely supervised by appropriate regulatory authorities. All studies that involve a medical or therapeutic intervention on patients must be approved by a supervising ethics committee before permission is granted to run the trial. The local ethics committee has discretion on how it will supervise noninterventional studies (observational studies or those using already collected data). In the U.S., this body is called the Institutional Review Board (IRB). Most IRBs are located at the local investigator's hospital or institution, but some sponsors allow the use of a central (independent/for profit) IRB for investigators who work at smaller institutions.
To be ethical, researchers must obtain the full and informed consent of participating human subjects. (One of the IRB's main functions is ensuring that potential patients are adequately informed about the clinical trial.) If the patient is unable to consent for him/herself, researchers can seek consent from the patient's legally authorized representative. In California, the state has prioritized the individuals who can serve as the legally authorized representative.
In some U.S. locations, the local IRB must certify researchers and their staff before they can conduct clinical trials. They must understand the federal patient privacy (HIPAA) law and good clinical practice. International Conference of Harmonisation Guidelines for Good Clinical Practice (ICH GCP) is a set of standards used internationally for the conduct of clinical trials. The guidelines aim to ensure that the "rights, safety and well being of trial subjects are protected".
The notion of informed consent of participating human subjects exists in many countries all over the world, but its precise definition may still vary.
Informed consent is clearly a necessary condition for ethical conduct but does not ensure ethical conduct. The final objective is to serve the community of patients or future patients in a best-possible and most responsible way. However, it may be hard to turn this objective into a well-defined quantified objective function. In some cases this can be done, however, as for instance for questions of when to stop sequential treatments (see Odds algorithm), and then quantified methods may play an important role.
Tuesday, March 24, 2009
SCOP
Nearly all proteins have structural similarities
with other proteins and, in many cases, share a
common evolutionary origin. The knowledge of
these relationships makes important contributions to
molecular biology and to other related areas of
science. It is central to our understanding of the
structure and evolution of proteins. It will play an
important role in the interpretation of the sequences
produced by the genome projects and, therefore, in
understanding the evolution of development.
The recent exponential growth in the number of
proteins whose structures have been determined by
X-ray crystallography and NMR spectroscopy
means that there is now a large and rapidly growing
corpus of information available. At present (January,
1995) the Brookhaven Protein Databank (PDB,
(Abola et al., 1987)) contains 3091 entries and the
number is increasing by about 100 a month. To
facilitate the understanding of, and access to, this
information, we have constructed the Structural
Classification of Proteins (scop) database. This
database provides a detailed and comprehensive
description of the structural and evolutionary
relationships of proteins whose three-dimensional
structures have been determined. It includes all proteins in the current version of the PDB and
almost all proteins for which structures have been
published but whose co-ordinates are not available
from the PDB.
The classification of protein structures in the
database is based on evolutionary relationships and
on the principles that govern their three-dimensional
structure. Early work on protein structures showed
that there are striking regularities in the ways in
which secondary structures are assembled (Levitt
& Chothia, 1976; Chothia et al., 1977) and in the
topologies of the polypeptide chains (Richardson,
1976, 1977; Sternberg & Thornton, 1976). These
regularities arise from the intrinsic physical and
chemical properties of proteins (Chothia, 1984;
Finkelstein&Ptitsyn, 1987) and provide the basis for
the classification of protein folds (Levitt & Chothia,
1976; Richardson, 1981). This early work has been
taken further inmore recent papers; see, for example,
Holm & Sander (1993), Orengo et al. (1993),
Overington et al. (1993) and Yee & Dill (1993). An
extensive bibliography of papers on the classification
and the determinants of protein folds is given in scop.
The method used to construct the protein
classification in scop is essentially the visual
inspection and comparison of structures though
various automatic tools are used to make the task
manageable and help provide generality. Given the current limitations of purely automatic procedures,
we believe this approach produces the most
accurate and useful results. The unit of classification
is usually the protein domain. Small
proteins, and most of those of medium size, have
a single domain and are, therefore, treated as a
whole. The domains in large proteins are usually
classified individually.
The classification is on hierarchical levels that
embody the evolutionary and structural relationships.
FAMILY. Proteins are clustered together into
families on the basis of one of two criteria that imply
their having a common evolutionary origin: first, all
proteins that have residue identities of 30% and
greater; second, proteins with lower sequence identities but whose functions and structures are
very similar; for example, globins with sequence
identities of 15%.
SUPERFAMILY. Families, whose proteins have
low sequence identities but whose structures and, in
many cases, functional features suggest that a
common evolutionary origin is probable, are placed
together in superfamilies; for example, actin, the
ATPase domain of the heat-shock protein and
hexokinase (Flaherty et al., 1991).
COMMONFOLD. Superfamilies and families are
defined as having a common fold if their proteins
have same major secondary structures in same
arrangement with the same topological connections.
In scop we give for each fold short descriptions of its
main structural features. Different proteins with the
same fold usually have peripheral elements of
secondary structure and turn regions that differ in
size and conformation and, in the more divergent
cases, these differing regions may form half or more
of each structure. For proteins placed together in the
same fold category, the structural similarities
probably arise from the physics and chemistry of
proteins favouring certain packing arrangements and
chain topologies (see above). There may, however,
be cases where a common evolutionary origin is
obscured by the extent of the divergence in sequence,
structure and function. In these cases, it is possible
that the discovery of new structures, with folds
between those of the previously known structures,
will make clear their common evolutionary relationship.
CLASS. For convenience of users, the different
folds have been grouped into classes. Most of the
folds are assigned to one of the five structural classes
on the basis of the secondary structures of which
they composed: (1) all alpha (for proteins whose
structure is essentially formed by a-helices), (2) all
beta (for those whose structure is essentially formed
by b-sheets), (3) alpha and beta (for proteins with
a-helices and b-strands that are largely interspersed),
(4) alpha plus beta (for those in which
a-helices and b-strands are largely segregated) and
(5)multi-domain (for those with domains of different
fold and for which no homologues are known at
present). Note that we do not use Greek characters
in scop because they are not accessible to all world
wide web viewers. More unusual proteins, peptides
and the PDB entries for designed proteins theoretical models, nucleic acids and carbohydrates,
have been assigned to other classes.
The number of entries, families, superfamilies and
common folds in the current version of scop are
shown in Figure 1. The exact position of boundaries
between family, superfamily and fold are, to some
degree, subjective. However, because all proteins
that could conceivably belong to a family or
superfamily are clustered together in the encompassing
fold category, some users may wish to
concentrate on this part of the database.
In addition to the information on structural and
evolutionary relationships, each entry (for which
co-ordinates are available) has links to images of the
structure, interactive molecular viewers, the atomic
co-ordinates, sequence data and homologues and
MEDLINE abstracts (see Table 1).
Two search facilities are available in scop. The
homology search permits users to enter a sequence
and obtain a list of any structures to which it has
significant levels of sequence similarity. The key
word search finds, for a word entered by the user,
matches from both the text of the scop database and
the headers of Brookhaven Protein Databank
structure files.
To provide easy and broad access, we have made
the scop database available as a set of tightly coupled
hypertext pages on the world wide web (WWW).
This allows it to be accessed by any machine on the
internet (including Macintoshes, PCs and workstations)
using freeWWWreader programs, such as
Mosaic (Schatz & Hardin, 1994). Once such a
program has been started, it is necessary only to
‘‘open’’ URL:
http://scop.mrc-lmb.cam.ac.uk/scop/
to obtain the ‘‘home’’ page level of the database.
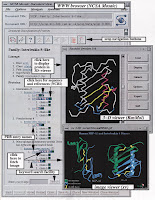
In Figure 2 we show a typical page from the
database. Each page has buttons to go back to the
top-level home page, to send electronic mail to the
authors, and to retrieve a detailed help page.
Navigating through the tree structure is simple;
selecting any entry retrieves the appropriate page. In
addition, buttons make it possible tomove within the
hierarchy in other manners, such as ‘‘upwards’’ to
obtain broader levels of classification.
The scop database was originally created as a
tool for understanding protein evolution through
sequence-structure relationships and determining if
new sequences and new structures are related to
previously known protein structures. On a more
general level, the highest levels of classification
provide an overview of the diversity of protein
structures now known and would be appropriate
both for researchers and students. The specific lower
levels should be helpful for comparing individual
structures with their evolutionary and structurally
related counterparts. In addition, we have also found
that the search capabilities with easy access to data
and images make scop a powerful general-purpose
interface to the PDB.
As new structures are released by PDB and
published, they will be entered in scop and revised versions of the database will be made available on
WWW. Moreover, as our formal understanding of
relationships between structure, sequence function
and evolution grows, it will be embodied in
additional facilities in the database.
Tuesday, March 17, 2009
XML (Extensible Markup Language)
XML (Extensible Markup Language) is a general-purpose specification for creating custom markup languages. It is classified as an extensible language, because it allows the user to define the mark-up elements. XML's purpose is to aid information systems in sharing structured data, especially via the Internet, to encode documents, and to serialize data; in the last context, it compares with text-based serialization languages such as JSON, YAML and S-Expressions.
XML's set of tools helps developers in creating web pages but its usefulness goes well beyond that. XML, in combination with other standards, makes it possible to define the content of a document separately from its formatting, making it easy to reuse that content in other applications or for other presentation environments. Most importantly, XML provides a basic syntax that can be used to share information between different kinds of computers, different applications, and different organizations without needing to pass through many layers of conversion.
XML began as a simplified subset of the Standard Generalized Markup Language (SGML), meant to be readable by people via semantic constraints; application languages can be implemented in XML. These include XHTML, RSS, MathML, GraphML, Scalable Vector Graphics, MusicXML, and others. Moreover, XML is sometimes used as the specification language for such application languages.
XML is recommended by the World Wide Web Consortium (W3C). It is a fee-free open standard. The recommendation specifies lexical grammar and parsing requirements.
Correctness
An XML document has two correctness levels:
· Well-formed. A well-formed document conforms to the XML syntax rules; e.g. if a start-tag (< >) appears without a corresponding end-tag (), it is not well-formed. A document not well-formed is not in XML; a conforming parser is disallowed from processing it.
· Valid. A valid document additionally conforms to semantic rules, either user-defined or in an XML schema, especially DTD; e.g. if a document contains an undefined element, then it is not valid; a validating parser is disallowed from processing it.
Well-formedness
If only a well-formed element is required, XML is a generic framework for storing any amount of text or any data whose structure can be represented as a tree. The only indispensable syntactical requirement is that the document has exactly one root element (also known as the document element), i.e. the text must be enclosed between a root start-tag and a corresponding end-tag, known as a "well-formed" XML document:
The root element can be preceded by an optional XML declaration element stating what XML version is in use (normally 1.0); it might also contain character encoding and external dependencies information.
The specification requires that processors of XML support the pan-Unicode character encodings UTF-8 and UTF-16 (UTF-32 is not mandatory). The use of more limited encodings, e.g. those based on ISO/IEC 8859, is acknowledged, widely used, and supported.
Comments can be placed anywhere in the tree, including in the text if the content of the element is text or #PCDATA.
XML comments start with . Two consecutive dashes (--) may not appear anywhere in the text of the comment.
In any meaningful application, additional markup is used to structure the contents of the XML document. The text enclosed by the root tags may contain an arbitrary number of XML elements. The basic syntax for one element is:
The two instances of »element_name« are referred to as the start-tag and end-tag, respectively. Here, »Element Content« is some text which may again contain XML elements. So, a generic XML document contains a tree-based data structure. Here is an example of a structured XML document:
Attribute values must always be quoted, using single or double quotes, and each attribute name may appear only once in any single element.
XML requires that elements be properly nested—elements may never overlap, and so must be closed in the order opposite to which they are opened. For example, this fragment of code below cannot be part of a well-formed XML document because the title and author elements are closed in the wrong order:
One way of writing the same information in a way which could be incorporated into a well-formed XML document is as follows:
XML provides special syntax for representing an element with empty content. Instead of writing a start-tag followed immediately by an end-tag, a document may contain an empty-element tag. An empty-element tag resembles a start-tag but contains a slash just before the closing angle bracket. The following three examples are equivalent in XML:
An empty-element may contain attributes:
Entity references
An entity in XML is a named body of data, usually text. Entities are often used to represent single characters that cannot easily be entered on the keyboard; they are also used to represent pieces of standard ("boilerplate") text that occur in many documents, especially if there is a need to allow such text to be changed in one place only.
Special characters can be represented either using entity references, or by means of numeric character references. An example of a numeric character reference is "€", which refers to the Euro symbol by means of its Unicode codepoint in hexadecimal.
An entity reference is a placeholder that represents that entity. It consists of the entity's name preceded by an ampersand ("&") and followed by a semicolon (";"). XML has five predeclared entities:
· & (& or "ampersand")
· < (< or "less than")
· > (> or "greater than")
· ' (' or "apostrophe")
· " (" or "quotation mark")
Here is an example using a predeclared XML entity to represent the ampersand in the name "AT&T":
Additional entities (beyond the predefined ones) can be declared in the document's Document Type Definition (DTD). A basic example of doing so in a minimal internal DTD follows. Declared entities can describe single characters or pieces of text, and can reference each other.
]>
©right-notice;
Numeric character references
Numeric character references look like entity references, but instead of a name, they contain the "#" character followed by a number. The number (in decimal or "x"-prefixed hexadecimal) represents a Unicode code point. Unlike entity references, they are neither predeclared nor do they need to be declared in the document's DTD. They have typically been used to represent characters that are not easily encodable, such as an Arabic character in a document produced on a European computer. The ampersand in the "AT&T" example could also be escaped like this (decimal 38 and hexadecimal 26 both represent the Unicode code point for the "&" character):
Similarly, in the previous example, notice that "©" is used to generate the “©” symbol.
See also numeric character references.
Well-formed documents
In XML, a well-formed document must conform to the following rules, among others:
· Non-empty elements are delimited by both a start-tag and an end-tag.
· Empty elements may be marked with an empty-element (self-closing) tag, such as
· All attribute values are quoted with either single (') or double (") quotes. Single quotes close a single quote and double quotes close a double quote.[citation needed]
· To include a double quote inside an attribute value that is double quoted, or a single quote inside an attribute value that is single quoted, escape the inner quote mark using a Character_entity_reference. This is necessary when an attribute value must contain both types (single and double quotes) or when you do not have control over the type of quotation a particular XML editor uses for wrapping attribute values. These character entity references are predefined in XML and do not need to be declared even when using a DTD or Schema: " and '. You may also use the numeric character entity references (hex) " and ' or their equivalent decimal notations " and '.
· Tags may be nested but must not overlap. Each non-root element must be completely contained in another element.
· The document complies with its declared character encoding. The encoding may be declared or implied externally, such as in "Content-Type" headers when a document is transported via HTTP, or internally, using explicit markup at the very beginning of the document. When no such declaration exists, a Unicode encoding is assumed, as defined by a Unicode Byte Order Mark before the document's first character. If the mark does not exist, UTF-8 encoding is assumed.
Element names are case-sensitive. For example, the following is a well-formed matching pair:
whereas these are not
By carefully choosing the names of the XML elements one may convey the meaning of the data in the markup. This increases human readability while retaining the rigor needed for software parsing.
Choosing meaningful names implies the semantics of elements and attributes to a human reader without reference to external documentation. However, this can lead to verbosity, which complicates authoring and increases file size.
Automatic verification
It is relatively simple to verify that a document is well-formed or validated XML, because the rules of well-formedness and validation of XML are designed for portability of tools. The idea is that any tool designed to work with XML files will be able to work with XML files written in any XML language (or XML application). Here are some examples of ways to verify XML documents:
· load it into an XML-capable browser, such as Firefox or Internet Explorer
· use a tool like xmlwf (usually bundled with expat)
· parse the document, for instance in Ruby:
irb> require "rexml/document"
irb> include REXML
irb> doc = Document.new(File.new("test.xml")).root
Validity
By leaving the names, allowable hierarchy, and meanings of the elements and attributes open and definable by a customizable schema or DTD, XML provides a syntactic foundation for the creation of purpose-specific, XML-based markup languages. The general syntax of such languages is rigid — documents must adhere to the general rules of XML, ensuring that all XML-aware software can at least read and understand the relative arrangement of information within them. The schema merely supplements the syntax rules with a set of constraints. Schemas typically restrict element and attribute names and their allowable containment hierarchies, such as only allowing an element named 'birthday' to contain one element named 'month' and one element named 'day', each of which has to contain only character data. The constraints in a schema may also include data type assignments that affect how information is processed; for example, the 'month' element's character data may be defined as being a month according to a particular schema language's conventions, perhaps meaning that it must not only be formatted a certain way, but also must not be processed as if it were some other type of data.
An XML document that complies with a particular schema/DTD, in addition to being well-formed, is said to be valid.
An XML schema is a description of a type of XML document, typically expressed in terms of constraints on the structure and content of documents of that type, above and beyond the basic constraints imposed by XML itself. A number of standard and proprietary XML schema languages have emerged for the purpose of formally expressing such schemas, and some of these languages are XML-based, themselves.
Before the advent of generalised data description languages such as SGML and XML, software designers had to define special file formats or small languages to share data between programs. This required writing detailed specifications and special-purpose parsers and writers.
XML's regular structure and strict parsing rules allow software designers to leave parsing to standard tools, and since XML provides a general, data model-oriented framework for the development of application-specific languages, software designers need only concentrate on the development of rules for their data, at relatively high levels of abstraction.
Well-tested tools exist to validate an XML document "against" a schema: the tool automatically verifies whether the document conforms to constraints expressed in the schema. Some of these validation tools are included in XML parsers, and some are packaged separately.
Other usages of schemas exist: XML editors, for instance, can use schemas to support the editing process (by suggesting valid elements and attributes names, etc).
DTD
The oldest schema format for XML is the Document Type Definition (DTD), inherited from SGML. While DTD support is ubiquitous due to its inclusion in the XML 1.0 standard, it is seen as limited for the following reasons:
· It has no support for newer features of XML, most importantly namespaces.
· It lacks expressiveness. Certain formal aspects of an XML document cannot be captured in a DTD.
· It uses a custom non-XML syntax, inherited from SGML, to describe the schema.
DTD is still used in many applications because it is considered the easiest to read and write.
XML Schema
A newer XML schema language, described by the W3C as the successor of DTDs, is XML Schema, or more informally referred to by the initialism for XML Schema instances, XSD (XML Schema Definition). XSDs are far more powerful than DTDs in describing XML languages. They use a rich datatyping system, allow for more detailed constraints on an XML document's logical structure, and must be processed in a more robust validation framework. XSDs also use an XML-based format, which makes it possible to use ordinary XML tools to help process them, although XSD implementations require much more than just the ability to read XML.
RELAX NG
Another popular schema language for XML is RELAX NG. Initially specified by OASIS, RELAX NG is now also an ISO international standard (as part of DSDL). It has two formats: an XML based syntax and a non-XML compact syntax. The compact syntax aims to increase readability and writability but, since there is a well-defined way to translate the compact syntax to the XML syntax and back again by means of James Clark's Trang conversion tool, the advantage of using standard XML tools is not lost. RELAX NG has a simpler definition and validation framework than XML Schema, making it easier to use and implement. It also has the ability to use datatype framework plug-ins; a RELAX NG schema author, for example, can require values in an XML document to conform to definitions in XML Schema Datatypes.
ISO DSDL and other schema languages
The ISO DSDL (Document Schema Description Languages) standard brings together a comprehensive set of small schema languages, each targeted at specific problems. DSDL includes RELAX NG full and compact syntax, Schematron assertion language, and languages for defining datatypes, character repertoire constraints, renaming and entity expansion, and namespace-based routing of document fragments to different validators. DSDL schema languages do not have the vendor support of XML Schemas yet, and are to some extent a grassroots reaction of industrial publishers to the lack of utility of XML Schemas for publishing.
Some schema languages not only describe the structure of a particular XML format but also offer limited facilities to influence processing of individual XML files that conform to this format. DTDs and XSDs both have this ability; they can for instance provide attribute defaults. RELAX NG and Schematron intentionally do not provide these; for example the infoset augmentation facility.
International use
XML supports the direct use of almost any Unicode character in element names, attributes, comments, character data, and processing instructions (other than the ones that have special symbolic meaning in XML itself, such as the open corner bracket, "<"). Therefore, the following is a well-formed XML document, even though it includes both Chinese and Cyrillic characters:
<俄語>Китайский
Displaying on the web
Generally, generic XML documents do not carry information about how to display the data.Without using CSS or XSLT, a generic XML document is rendered as raw XML text by most web browsers. Some display it with 'handles' (e.g. + and - signs in the margin) that allow parts of the structure to be expanded or collapsed with mouse-clicks.
In order to style the rendering in a browser with CSS, the XML document must include a reference to the stylesheet:
Note that this is different from specifying such a stylesheet in HTML, which uses the element.
XSLT (XSL Transformations) can be used to alter the format of XML data, either into HTML or other formats that are suitable for a browser to display.
To specify client-side XSLT, the following processing instruction is required in the XML:
Client-side XSLT is supported by many web browsers. Alternatively, one may use XSLT to convert XML into a displayable format on the server rather than being dependent on the end-user's browser capabilities. The end-user is not aware of what has gone on 'behind the scenes'; all they see is well-formatted, displayable data.
See the XSLT article for examples of XSLT in action.
Extensions
· XPath makes it possible to refer to individual parts of an XML document. This provides random access to XML data for other technologies, including XSLT, XSL-FO, XQuery etc. XPath expressions can refer to all or part of the text, data and values in XML elements, attributes, processing instructions, comments etc. They can also access the names of elements and attributes. XPaths can be used in both valid and well-formed XML, with and without defined namespaces.
· XInclude defines the ability for XML files to include all or part of an external file. When processing is complete, the final XML infoset has no XInclude elements, but instead has copied the documents or parts thereof into the final infoset. It uses XPath to refer to a portion of the document for partial inclusions.
· XQuery is to XML and XML Databases what SQL and PL/SQL are to relational databases: ways to access, manipulate and return XML.
· XML Namespaces enable the same document to contain XML elements and attributes taken from different vocabularies, without any naming collisions occurring.
· XML Signature defines the syntax and processing rules for creating digital signatures on XML content.
· XML Encryption defines the syntax and processing rules for encrypting XML content.
· XPointer is a system for addressing components of XML-based internet media.
XML files may be served with a variety of Media types. RFC 3023 defines the types "application/xml" and "text/xml", which say only that the data is in XML, and nothing about its semantics. The use of "text/xml" has been criticized as a potential source of encoding problems but is now in the process of being deprecated. RFC 3023 also recommends that XML-based languages be given media types beginning in "application/" and ending in "+xml"; for example "application/atom+xml" for Atom. This page discusses further XML and MIME.
JDBC
Java Database Connectivity (JDBC) is an API for the Java programming language that defines how a client may access a database. It provides methods for querying and updating data in a database. JDBC is oriented towards relational databases.
The Java 2 Platform, Standard Edition, version 1.4 (J2SE) includes the JDBC 3.0 API together with a reference implementation JDBC-to-ODBC bridge, enabling connections to any ODBC-accessible data source in the JVM host environment. This bridge is native code (not Java), closed source, and only appropriate for experimental use and for situations in which no other driver is available, not least because it provides only a limited subset of the JDBC 3.0 API, as it was originally built and shipped with JDBC 1.0 for use with old ODBC v2.0 drivers (ODBC v3.0 was released in 1996).
Overview
JDBC has been part of the Java Standard Edition since the release of JDK 1.1. The JDBC classes are contained in the Java package java.sql. Starting with version 3.0, JDBC has been developed under the Java Community Process. JSR 54 specifies JDBC 3.0 (included in J2SE 1.4), JSR 114 specifies the JDBC Rowset additions, and JSR 221 is the specification of JDBC 4.0 (included in Java SE 6).
JDBC allows multiple implementations to exist and be used by the same application. The API provides a mechanism for dynamically loading the correct Java packages and registering them with the JDBC Driver Manager. The Driver Manager is used as a connection factory for creating JDBC connections.
JDBC connections support creating and executing statements. These may be update statements such as SQL's CREATE, INSERT, UPDATE and DELETE, or they may be query statements such as SELECT. Additionally, stored procedures may be invoked through a JDBC connection. JDBC represents statements using one of the following classes:
· Statement – the statement is sent to the database server each and every time.
· PreparedStatement – the statement is cached and then the execution path is pre determined on the database server allowing it to be executed multiple times in an efficient manner.
· CallableStatement – used for executing stored procedures on the database.
Update statements such as INSERT, UPDATE and DELETE return an update count that indicates how many rows were affected in the database. These statements do not return any other information.
Query statements return a JDBC row result set. The row result set is used to walk over the result set. Individual columns in a row are retrieved either by name or by column number. There may be any number of rows in the result set. The row result set has metadata that describes the names of the columns and their types.
There is an extension to the basic JDBC API in the javax.sql.
Example
The method Class.forName(String) is used to load the JDBC driver class. The line below causes the JDBC driver from some jdbc vendor to be loaded into the application. (Some JVMs also require the class to be instantiated with .newInstance().)
Class.forName( "com.somejdbcvendor.TheirJdbcDriver" );
In JDBC 4.0, it's no longer necessary to explicitly load JDBC drivers using Class.forName(). See JDBC 4.0 Enhancements in Java SE 6.
When a Driver class is loaded, it creates an instance of itself and registers it with the DriverManager. This can be done by including the needed code in the driver class's static block. e.g. DriverManager.registerDriver(Driver driver)
Now when a connection is needed, one of the DriverManager.getConnection() methods is used to create a JDBC connection.
Connection conn = DriverManager.getConnection(
"jdbc:somejdbcvendor:other data needed by some jdbc vendor",
"myLogin",
"myPassword" );
The URL used is dependent upon the particular JDBC driver. It will always begin with the "jdbc:" protocol, but the rest is up to the particular vendor. Once a connection is established, a statement must be created.
Statement stmt = conn.createStatement();
try {
stmt.executeUpdate( "INSERT INTO MyTable( name ) VALUES ( 'my name' ) " );
} finally {
//It's important to close the statement when you are done with it
stmt.close();
}
Note that Connections, Statements, and ResultSets often tie up operating system resources such as sockets or file descriptors. In the case of Connections to remote database servers, further resources are tied up on the server, e.g., cursors for currently open ResultSets. It is vital to close() any JDBC object as soon as it has played its part; garbage collection should not be relied upon. Forgetting to close() things properly results in spurious errors and misbehaviour. The above try-finally construct is a recommended code pattern to use with JDBC objects.
Data is retrieved from the database using a database query mechanism. The example below shows creating a statement and executing a query.
Statement stmt = conn.createStatement();
try {
ResultSet rs = stmt.executeQuery( "SELECT * FROM MyTable" );
try {
while ( rs.next() ) {
int numColumns = rs.getMetaData().getColumnCount();
for ( int i = 1 ; i <= numColumns ; i++ ) {
// Column numbers start at 1.
// Also there are many methods on the result set to return
// the column as a particular type. Refer to the Sun documentation
// for the list of valid conversions.
System.out.println( "COLUMN " + i + " = " + rs.getObject(i) );
}
}
} finally {
rs.close();
}
} finally {
stmt.close();
}
Typically, however, it would be rare for a seasoned Java programmer to code in such a fashion. The usual practice would be to abstract the database logic into an entirely different class and to pass preprocessed strings (perhaps derived themselves from a further abstracted class) containing SQL statements and the connection to the required methods. Abstracting the data model from the application code makes it more likely that changes to the application and data model can be made independently.
An example of a PreparedStatement query. Using conn and class from first example.
PreparedStatement ps = conn.prepareStatement( "SELECT i.*, j.* FROM Omega i, Zappa j WHERE i.name = ? AND j.num = ?" );
try {
// In the SQL statement being prepared, each question mark is a placeholder
// that must be replaced with a value you provide through a "set" method invocation.
// The following two method calls replace the two placeholders; the first is
// replaced by a string value, and the second by an integer value.
ps.setString(1, "Poor Yorick");
ps.setInt(2, 8008);
// The ResultSet, rs, conveys the result of executing the SQL statement.
// Each time you call rs.next(), an internal row pointer, or cursor,
// is advanced to the next row of the result. The cursor initially is
// positioned before the first row.
ResultSet rs = ps.executeQuery();
try {
while ( rs.next() ) {
int numColumns = rs.getMetaData().getColumnCount();
for ( int i = 1 ; i <= numColumns ; i++ ) {
// Column numbers start at 1.
// Also there are many methods on the result set to return
// the column as a particular type. Refer to the Sun documentation
// for the list of valid conversions.
System.out.println( "COLUMN " + i + " = " + rs.getObject(i) );
} // for
} // while
} finally {
rs.close();
}
} finally {
ps.close();
} // try
A typical implementation model of Java-RMI using stub and skeleton objects. Java 2 SDK, Standard Edition, v1.2 removed the need for a skeleton.
The Java Remote Method Invocation API, or Java RMI, a Java application programming interface, performs the object-oriented equivalent of remote procedure calls.
Two common implementations of the API exist:
1. The original implementation depends on Java Virtual Machine (JVM) class representation mechanisms and it thus only supports making calls from one JVM to another. The protocol underlying this Java-only implementation is known as Java Remote Method Protocol (JRMP).
2. In order to support code running in a non-JVM context, a CORBA version was later developed.
Usage of the term RMI may denote solely the programming interface or may signify both the API and JRMP, whereas the term RMI-IIOP (read: RMI over IIOP) denotes the RMI interface delegating most of the functionality to the supporting CORBA implementation.
The programmers of the original RMI API generalized the code somewhat to support different implementations, such as an HTTP transport. Additionally, work was done to CORBA, adding a pass-by-value capability, to support the RMI interface. Still, the RMI-IIOP and JRMP implementations do not have fully identical interfaces.
RMI functionality comes in the package java.rmi, while most of Sun's implementation is located in the sun.rmi package. Note that with Java versions before Java 5.0 developers had to compile RMI stubs in a separate compilation step using rmic. Version 5.0 of Java and beyond no longer require this step.Jini offers a more advanced version of RMI in Java - it functions similarly but provides more advanced searching capabilities and mechanisms for distributed object applications.
J2EE
Introduction to J2EE
Java Platform, Enterprise Edition Version 2 or J2EE is a widely used platform for server programming in the Java programming language. The Java EE Platform differs from the Java Standard Edition Platform (Java SE) in that it adds libraries which provide functionality to deploy fault-tolerant, distributed, multi-tier Java software, based largely on modular components running on an application server.
Nomenclature, standards and specifications
The platform was known as Java 2 Platform, Enterprise Edition or J2EE until the name was changed to Java EE in version 5. The current version is called Java EE 5. The previous version is called J2EE 1.4.
Java EE is defined by its specification. As with other Java Community Process specifications, Java EE is also considered informally to be a standard since providers must agree to certain conformance requirements in order to declare their products as Java EE compliant; albeit with no ISO or ECMA standard.
Java EE includes several API specifications, such as JDBC, RMI, e-mail, JMS, web services, XML, etc, and defines how to coordinate them. Java EE also features some specifications unique to Java EE for components. These include Enterprise JavaBeans, servlets, portlets (following the Java Portlet specification), JavaServer Pages and several web service technologies. This allows developers to create enterprise applications that are portable and scalable, and that integrate with legacy technologies. A Java EE "application server" can handle the transactions, security, scalability, concurrency and management of the components that are deployed to it, meaning that the developers should be able to concentrate more on the business logic of the components rather than on infrastructure and integration tasks.
History
The original J2EE specification was developed by Sun Microsystems.
The J2EE 1.2 SDK was released in December 1999. Starting with J2EE 1.3, the specification was developed under the Java Community Process. Java Specification Request (JSR) 58 specifies J2EE 1.3 and JSR 151 specifies the J2EE 1.4 specification.The J2EE 1.3 SDK was first released by Sun as a beta in April 2001. The J2EE 1.4 SDK beta was released by Sun in December 2002.The Java EE 5 specification was developed under JSR 244 and the final release was made on May 11, 2006.
The Java EE 6 specification has been developed under JSR 316 and is scheduled for release in May, 2009.
General APIs
The Java EE APIs includes several technologies that extend the functionality of the base Java SE APIs.
javax.ejb.*
The Enterprise JavaBean's 1st and 2nd API defines a set of APIs that a distributed object container will support in order to provide persistence, remote procedure calls (using RMI or RMI-IIOP), concurrency control, and access control for distributed objects. This package contains the Enterprise JavaBeans classes and interfaces that define the contracts between the enterprise bean and its clients and between the enterprise bean and the ejb container. This package contains the maximum number of Exception classes (16 in all) in Java EE 5 SDK.
javax.transaction.*
These packages define the Java Transaction API (JTA).
javax.xml.stream
This package contains readers and writers for XML streams. This package contains the only Error class in Java EE 5 SDK.
javax.jms.*
This package defines the Java Message Service (JMS) API. The JMS API provides a common way for Java programs to create, send, receive and read an enterprise messaging system's messages. This package has the maximum number of interfaces (43 in all) in the Java EE 5 SDK.
javax.faces.component.html
This package defines the JavaServer Faces (JSF) API. JSF is a technology for constructing user interfaces out of components.
javax.persistence
This package contains the classes and interfaces that define the contracts between a persistence provider and the managed classes and the clients of the Java Persistence API. This package contains the maximum number of annotation types (64 in all) and enums (10 in all) in the Java EE 5 SDK.
Certified application servers
Java EE 5 certified
· Sun Java System Application Server Platform Edition 9.0, based on the open-source server GlassFish
· GlassFish
· JBoss Application Server Version 5 [1] [2]
· Apache Geronimo 2.0
· Apache OpenEJB via Apache Geronimo
· IBM WebSphere Application Server Community Edition 2.0, based on Apache Geronimo
· IBM WebSphere Application Server V7
· WebLogic Application Server 10.0 from BEA Systems
· Oracle Containers for Java EE 11
· SAP NetWeaver Application Server, Java EE 5 Edition from SAP
· JEUS 6, an Application Server from TmaxSoft
J2EE 1.4 certified
· JBoss 4.x, an open-source application server from JBoss.
· Apache Geronimo 1.0, an open-source application server
· Pramati Server 5.0
· JOnAS, an open-source application server from ObjectWeb
· Oracle Application Server 10g
· Resin, an application server with integrated XML support
· SAP NetWeaver Application Server, Java EE 5 Edition from SAP AG
· Sun Java System Web Server
· Sun Java System Application Server Platform Edition 8.2
· IBM WebSphere Application Server (WAS)
· BEA Systems WebLogic server 8
· JEUS 5 from TmaxSoft